Klebsiella data visualisations
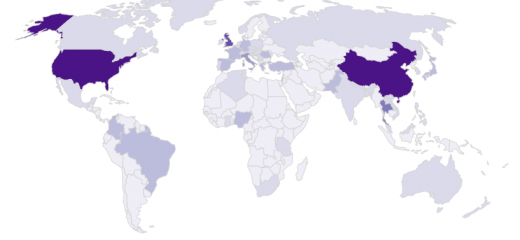
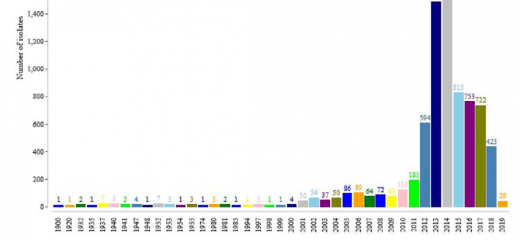
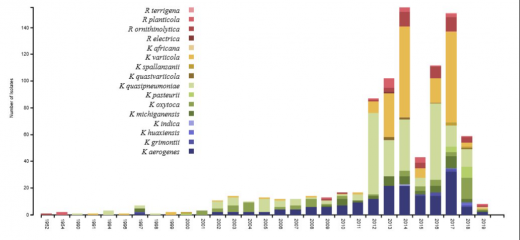
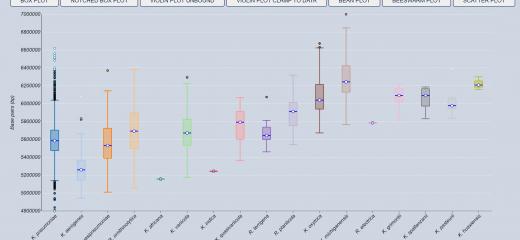
For the bacterial species validation we used Klebsiella isolates obtained from the most frequently used large genome databases (NCBI and ENA-SRA). These represent a collection of all Klebsiella isolates that have been cultured and sequenced historically (to the 8th July 2020). You can explore the different features of these data, putting the findings of the species validation story into context with their provenance.
It is important to note that these data are not a systematically collected set of bacterial isolates. They represent Klebsiella isolated from a variety of sources including: the environment, plants, animals and humans. Additional metadata describing their provenance (year/region of isolation) is not always provided by the scientists who upload the genome data to public repositories.