Klebsiella species: scalable genomic species assignment and validation
What are Klebsiella species?
-
Defining bacterial species is a complex task, involving assessments of phenotypic and genetic characteristics unique to a set of bacteria and differ sufficiently from other bacterial species.
-
Klebsiella spp. are a large group of bacteria, found ubiquitously in the environment and growing on and in plants, animals and humans (see Klebsiella "Explainers", section Klebsiella species information). The study of Klebsiella spp. is vitally important as they are a cause and contributor to the global problem of antimicrobial resistance (AMR).
-
Many important species within the Klebsiella genus are of interest to scientists and clinicians worldwide. Therefore, they need to accurately distinguish the 13 Klebsiella spp. recognised to date and the 4 closely-related Raoultella spp.
-
Determining species within the Klebsiella genus is difficult using traditional laboratory methods. Over time, many isolates have been named using these techniques. These isolates are now being recharacterized using high-resolution genomics methods, which involves studying bacterial DNA (see Klebsiella "Explainers", section The challenges with determining Klebsiella species).
-
Genomics is the only way to reliably speciate Klebsiella, as the different species have overlapping phenotypes and are genetically very closely related.
There is plenty of additional information, details and references available for this story in the Klebsiella "Explainers" section. So click here to browse this information or click on the links in the text below.
Aim
- To demonstrate Ribosomal Multilocus Sequence Typing (rMLST), as a tool to perform scalable, rapid and accurate species annotation.
- To validate the species of 10,570 Klebsiella isolates using rMLST and 17 type strain isolates and corroborate these findings with existing species identification methods.
Dataset
The PubMLST Multi-species isolate database contains isolate data and associated genome sequences obtained from two places:
- NCBI Assembly database
- Genome assembled in-house from data at the ENA Sequence Read Archive (ENA-SRA)
A dataset of 10,587 Klebsiella/Raoultella isolates was identified in the PubMLST Multi-species database (8th July 2020) and divided into 10,570 ‘query’ isolates and 17 type strain isolates. These isolates are publicly available on the PubMLST Multi-species website (go to the Klebsiella "Explainers" section to find out more about this website).
Type strain identification
To determine species, we needed to define a frame of reference to compare the unknown isolates to and we call these type strains. We identified 17 type strain isolates by examining NCBI Assembly information and cross-referencing this with the defined species for Klebsiella/Raoultella at NCBI Taxonomy and references therein. A table of these type strain isolates is found here.
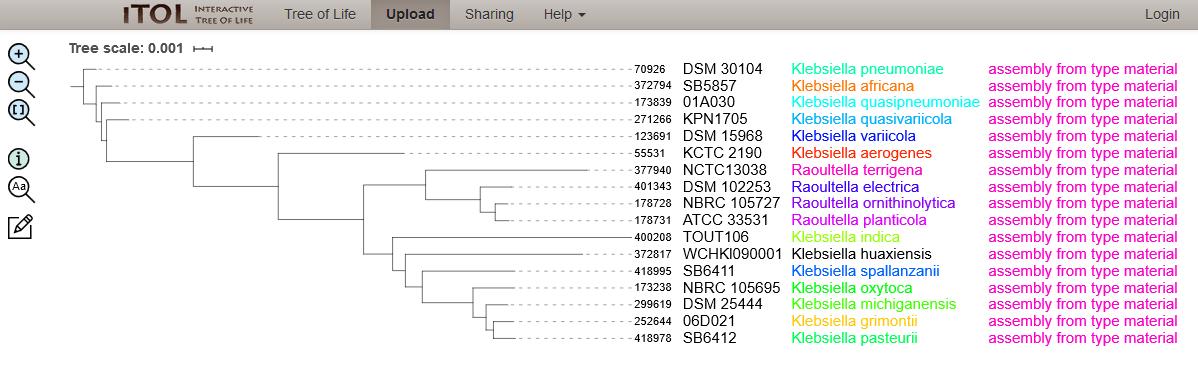
Figure 1: Phylogenetic tree of 17 Klebsiella/Raoultella type strain isolates. The neighbour-joining tree was calculated from the concatenated alleles of 51 rMLST loci and visualised using the Interactive Tree of Life (iTOL) website.
Automated genomic methods used to analyse Klebsiella species annotations
The species validation process is based on the underlying premise that the isolate has the same species as the nearest type strain isolate as measured by a nucleotide identity-based metric. Figure 2 shows an example of the rMLST allele-based phylogenetic tree of 17 Klebsiella/Raoultella type strain isolates and 15 additional Klebsiella aerogenes isolates. It is possible to see that the K. aerogenes isolates cluster very closely to the K. aerogenes type strain (KCTC 2190).
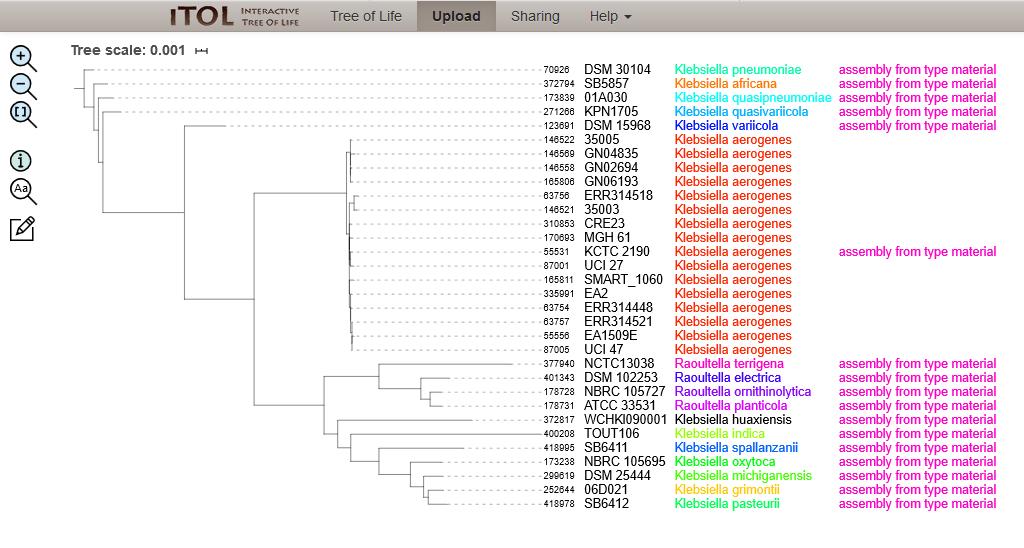
Figure 2: Phylogenetic tree of 17 Klebsiella/Raoultella type strain isolates and 15 additional Klebsiella aerogenes isolates. The neighbour joining tree was calculated from the concatenated alleles of 51 rMLST loci and visualised using the Interactive Tree of Life (iTOL) website.
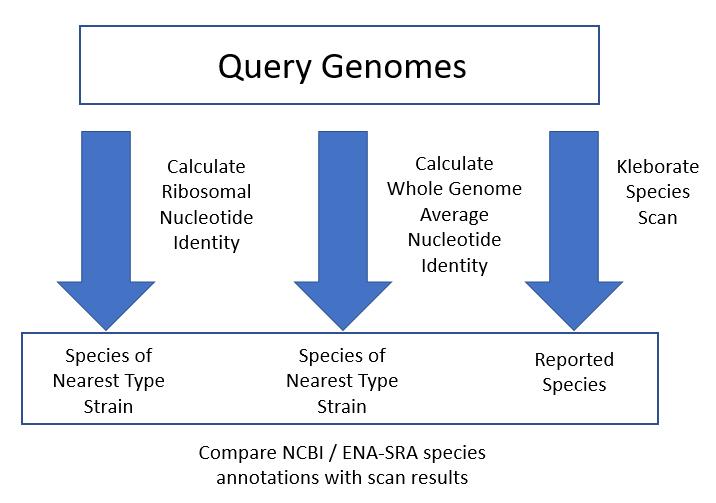
Figure 3: Three automated methods were used to analyse the species annotations of Klebsiella and Raoultella isolates. rMLST Ribosomal Nucleotide Identity, wgANI and the Kleborate species scanner were applied to the query genomes and the results were compared with the species annotation obtained from the source database.
We compared three automated methods of DNA comparison across the Klebsiella/Raoultella query dataset (Figure 3), (see Klebsiella "Explainers", section Genomic methods).
Overview of consistent species identification
There were 10,176/10,570 (96.3%) Klebsiella/Raoultella isolates from NCBI and ENA-SRA that were found to have consistent species annotations with the source database across all three automated species identification methods (rMLST Ribosomal Nucleotide Identity, wgANI and Kleborate species scan). All 10,176 isolates species annotations were confirmed by visual inspection on a phylogenetic tree of the 17 type strains. The table shows the number of isolates with validated species annotations per species.
| Species | Number of query isolates with consistent species annotation (number of type strains used) |
|---|---|
| Klebsiella aerogenes | 237 (1) |
| Klebsiella africana | 0 (1) |
| Klebsiella grimontii | 7 (1) |
| Klebsiella huaxiensis | 2 (1) |
| Klebsiella indica | 0 (1) |
| Klebsiella michiganensis | 72 (1) |
| Klebsiella oxytoca | 109 (1) |
| Klebsiella pasteurii | 12 (1) |
| Klebsiella pneumoniae | 9,047 (1) |
| Klebsiella quasipneumoniae | 298 (1) |
| Klebsiella quasivariicola | 5 (1) |
| Klebsiella spallanzanii | 3 (1) |
| Klebsiella variicola | 286 (1) |
| Raoultella electrica | 0 (1) |
| Raoultella ornithinolytica | 61 (1) |
| Raoultella planticola | 28 (1) |
| Raoultella terrigena | 9 (1) |
| Total | 10,176 (17) |
Inconsistent species annotations
There were 394 NCBI Assembly entries with a species annotation that was inconsistent, identified by all three automated methods.
| Species | No. of isolates with inconsistent species identification |
|---|---|
| Klebsiella species mismatch | 371 |
| Raoultella species mismatch | 6 |
| Labelled as Klebsiella species but matched Raoultella type strain | 4 |
| Not closely related to any Klebsiella/Raoultella type strains | 11 |
These 394 Klebsiella/Raoultella entries in the PubMLST Multi-species database have been removed from public view to avoid confusion. The isolates are retained in the database so that if the species annotations are updated by the source database, they can be re-analysed and made public as required. The NCBI Assembly database curators review species annotations based on contributor feedback and it is hoped that these entries will be updated in due course.
Example of an inconsistent species annotation
The NCBI Assembly entry for GCA_900083755.1 (Strain 2880STDY5682802) is annotated as Klebsiella oxytoca (as of 8th July 2020, Figure 4). Phylogenetic tree analysis based on rMLST alleles of the 17 type strain isolates and Strain 2880STDY5682802 shows that the query genome clusters with the type strain for Raoultella planticola (ATCC 33531) and is therefore considered to be an inconsistent species annotation (Figure 5). The whole genome ANI is 99.38% to ATCC 33531.
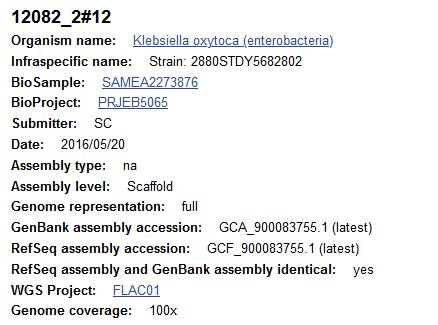
Figure 4: Screenshot of NCBI Assembly database entry for Strain: 2880STDY5682802 (GCA_900083755.1) on 8th July 2020 showing the species annotation of this isolate as Klebsiella oxytoca.
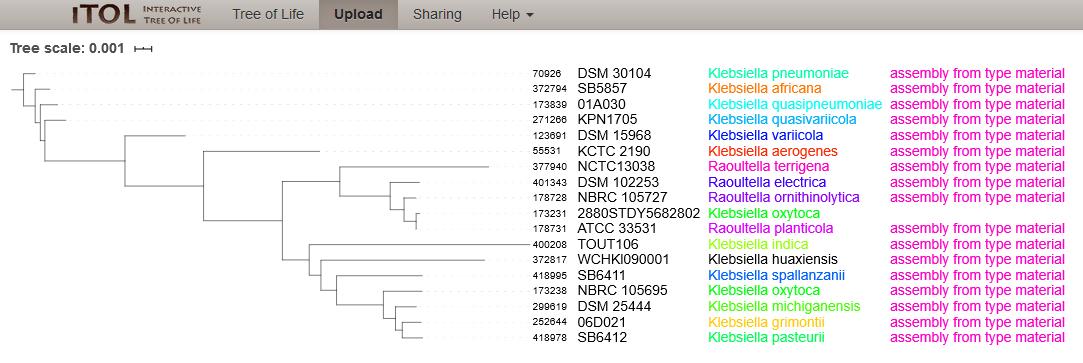
Figure 5: Phylogenetic tree of 17 type strain isolates in the Klebsiella/Raoultella genera with one entry from NCBI Assembly, Strain: 2880STDY5682802 (ID:173231) annotated as Klebsiella oxytoca (as of 8th July 2020). The tree was calculated from the concatenated alleles of 51 rMLST loci. The isolate clusters with the type strain for Raoultella planticola.
What have we learned?
- All three automated approaches gave consistent species annotation results for 10,570 Klebsiella isolates using WGS data.
- Some of the genomes in the NCBI Assembly database have been mis-assigned to species (394/10,570, 3.7%).
- The rMLST approach is both accurate in terms of species assignment and rapid compared to whole genome ANI. For more information see Klebsiella "Explainers", sections: What is rMLST? Phylogenetic analysis with rMLST and Species identifier.
- The species assignments made can be used to analyse isolates across the genus.
- rMLST is a multi-species approach and can be accessed via the rMLST database (new user log in required).
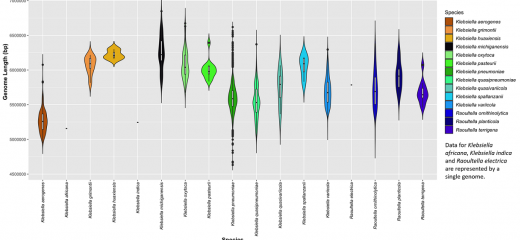
Accurately assigning species within a genus, such as Klebsiella, enables accurate genus-wide and species-specific analyses to be undertaken. For example the genome sizes of the isolates assigned for different species illustrate just how variable genome content can be even within members of the same species.
Use our interactive visualisation tools to explore and learn about the data