What can genomes do for us?
How do we use genome data to detect, identify and differentiate gonococci?
Genomes consist of DNA, the genetic blueprint of any living organism. Genomes from several thousand N. gonorrhoeae specimens can be compared and such studies allow us to understand what makes a gonococcus behave in certain ways. For example, we can find out why it may be resistant to antibiotic treatment.
Comparison of gonococcal genomes obtained worldwide also allows us to see if the same gonococci are found everywhere or if there are geographical differences.
The availability of genomes therefore offers us tantalising opportunities to forensically examine the gonococcus.
Image
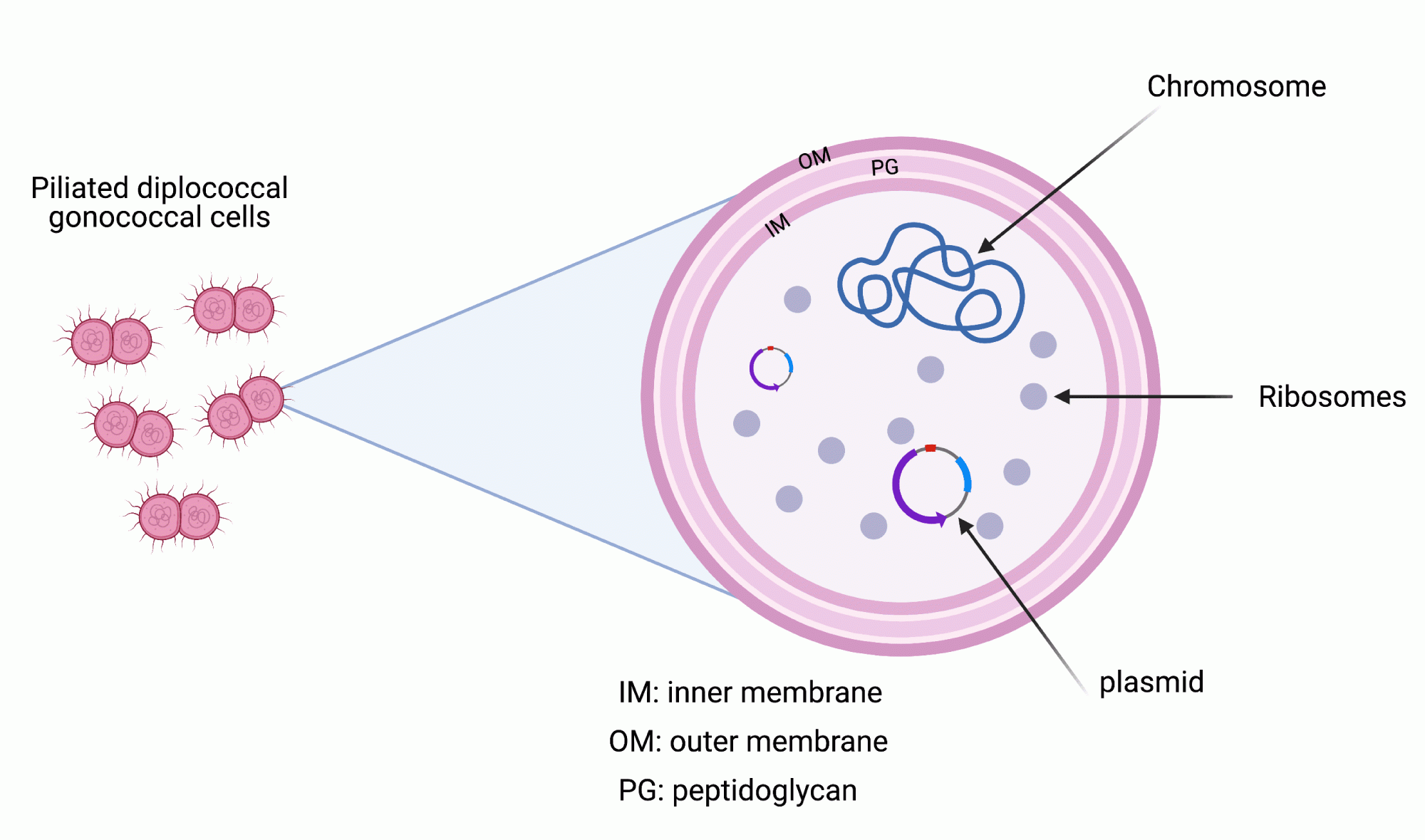
The gonococcus has a genome consisting of a single DNA molecule called a chromosome. It is just over 2 million base pairs in size and is circular. Some gonococci also possess extra chromosomal DNA known as plasmids which for the most part, confer resistance to antibiotics.
Over 2000 genes are encoded on the chromosome with around 1600 of these in common to all gonococci. These form part of the core genome.
Each gene has its own unique signature nucleotide sequence however genetic mutations arising from synonymous and/or non-synonymous nucleotide substitutions or genetic exchange (known as horizontal gene transfer) frequently occur generating new gene variants known as alleles.
Isolates will possess distinct combinations of allelic profiles.
Image
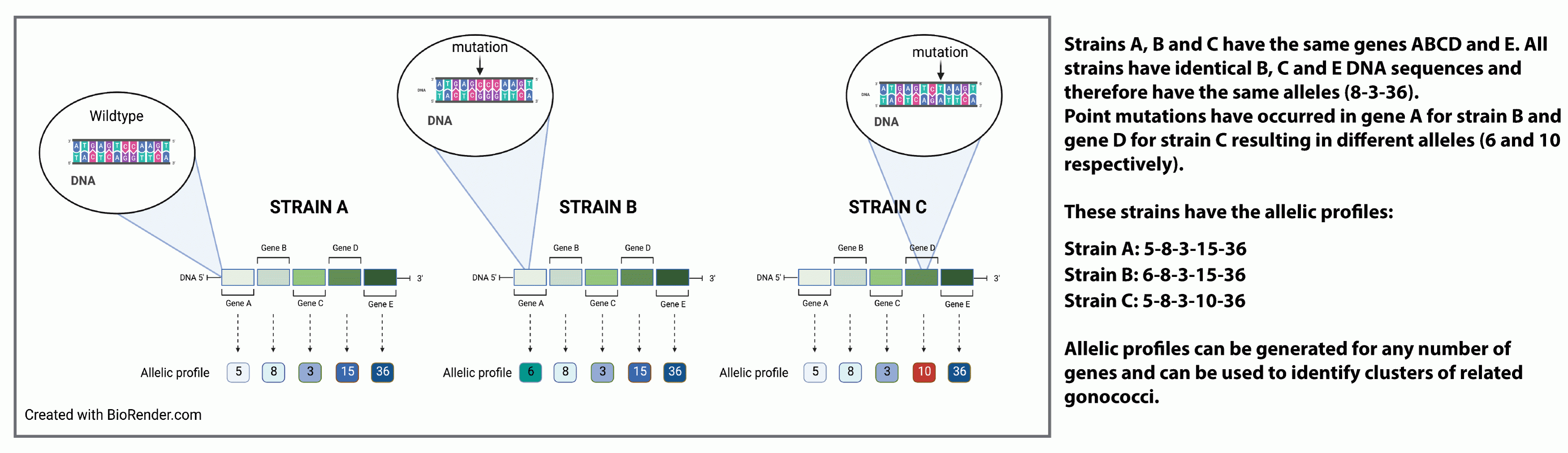
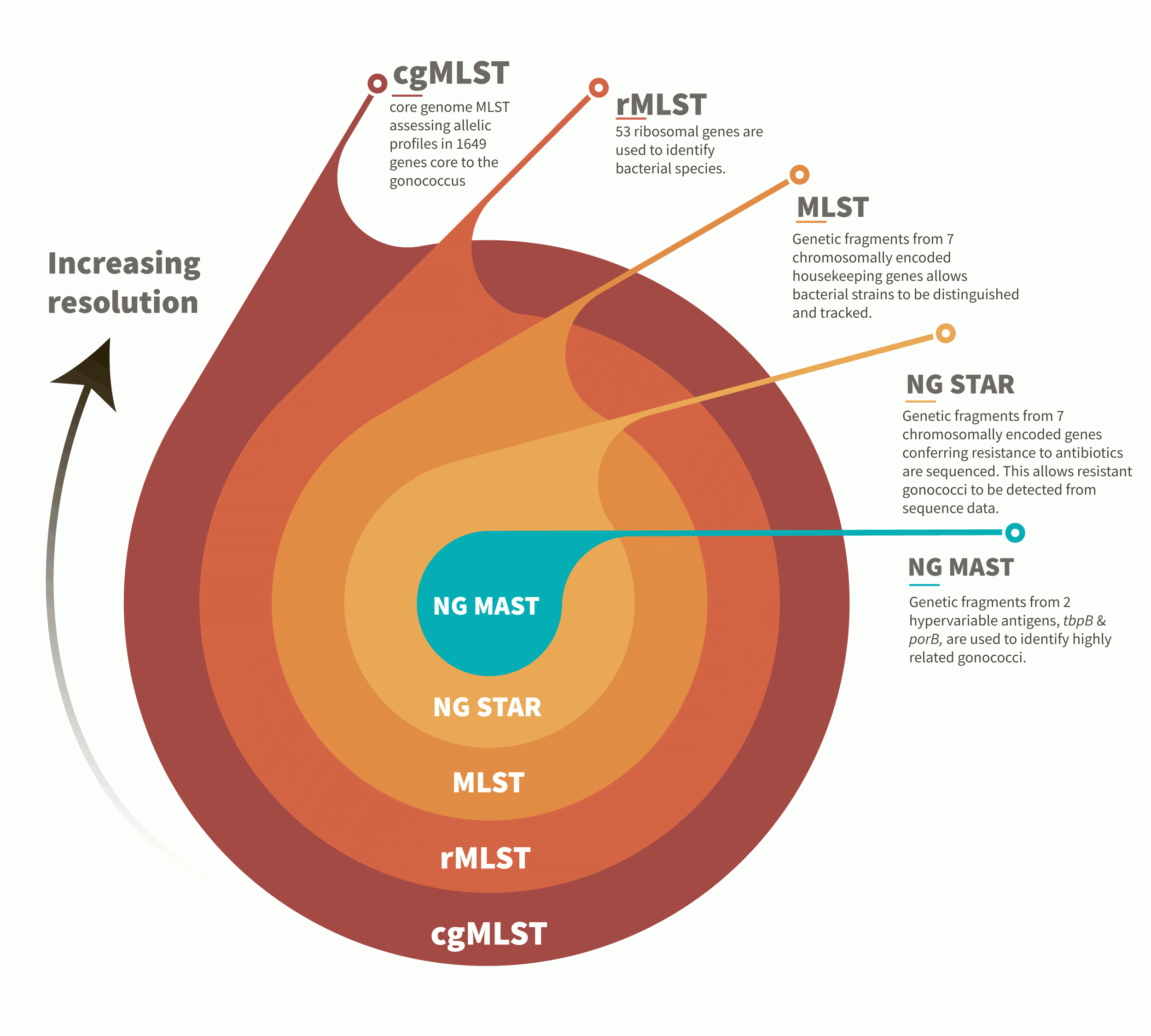
Allelic profiles can be organised into schemes consisting of multiple genes also known as loci (singular locus).
These are designated as multi-locus-sequence type, MLST.
Several MLST schemes have been developed for gonococcal studies:
•NG-MAST: where nucleotide sequences from fragments of porB and tbpB genes are used to define NG-MAST sequence types;
•NG STAR: (Neisseria gonorrhoeae Sequence Typing for Antimicrobial Resistance): which indexes variability found in 7 genes associated with AMR.
•MLST ST: which indexes variation found in 7 house-keeping gene fragments;
Gonococci can also be distinguished using ribosomal genes, rMLST, as well as the core genome, cgMLST.
Increasing numbers of loci compared improves the resolution obtained and allows us to find structure in the gonococcal population and identify clusters of related isolates.
Image
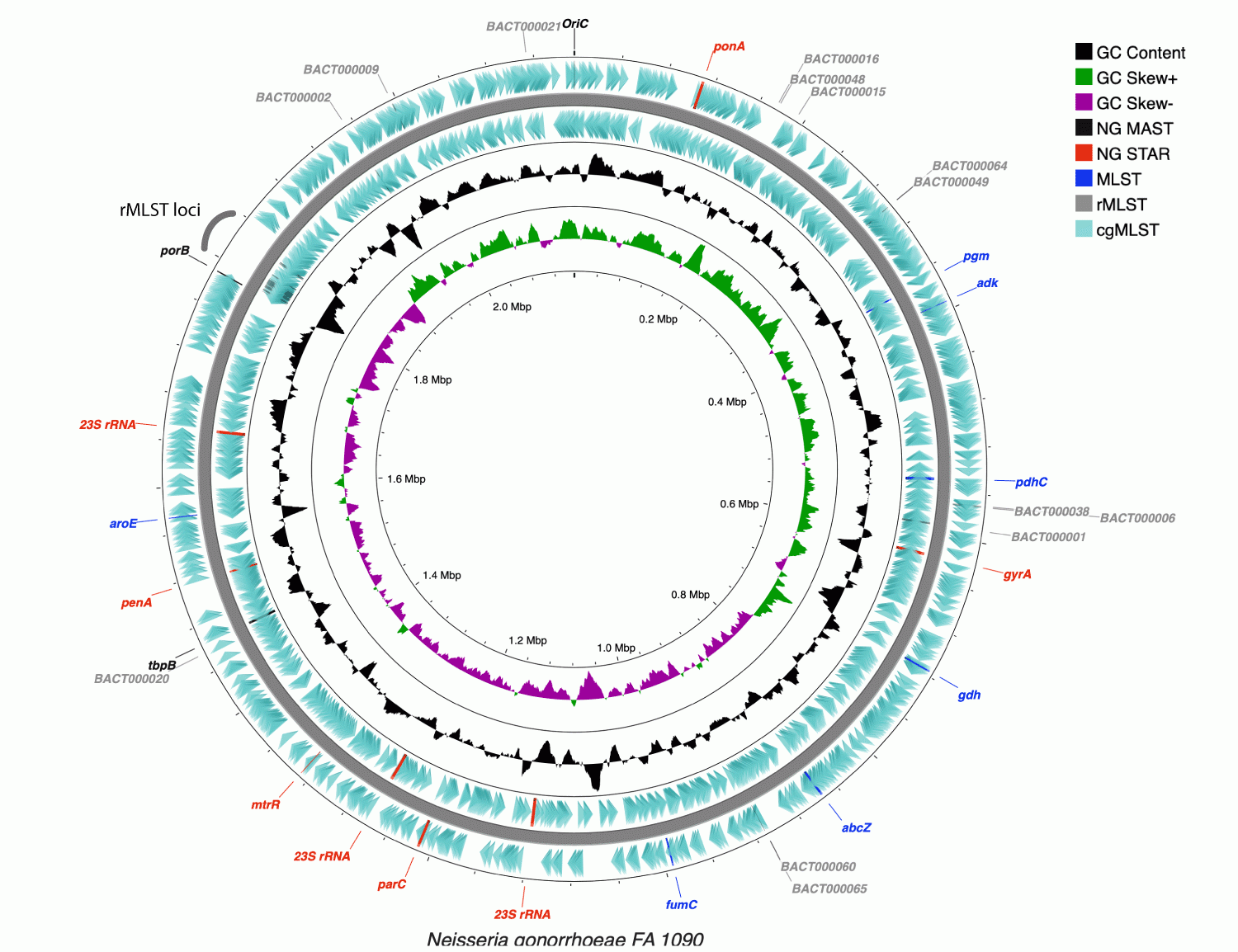
Schematic representation of the circular chromosome from the gonococcal strain, FA1090. On this the locations of genes associated with the NG MAST, NG STAR, MLST, rMLST and cgMLST schemes are indicated. Diagram generated using cgViewer
Studies examining the genetic diversity of gonococcal housekeeping genes have shown that, in addition to diversification arising from randomly distributed point mutations, these genes are frequently subject to exchange between gonococci. As a result, some gonococci, although possessing the same 7 locus MLST-ST, may have different ancestry at the genome level indicating that MLST STs cannot be used to reliably evaluate gonococcal populations.
Alternative approaches to the analysis of gonococcal populations therefore need to be employed. This includes the use of cgMLST where over 1600 genes can be used to distinguish gonococcal populations.
Typing schemes for the gonococcus begin with the NG MAST typing scheme which indexes diversity found at 2 hyper variable antigens allowing highly related clones to be identified.
NG STAR examines 7 loci known to confer resistance allowing chromosomal-mediated antimicrobial resistance to be deduced from genetic data.
MLST denotes allelic variation in 7 housekeeping genes and was the first molecular typing scheme used to differentiate the closely related bacterium, Neisseria meningitidis. See here for more about the meningococcus.
The ribosomal MLST scheme examines 53 genes essential to the ribosome and present across bacterial genera. See our pages on Klebsiella to find out more about how rMLST can be used.
Finally, the core genome, cgMLST examines genes core ie present in >95% of the gonococcal population. Using increasing numbers of genes in comparative analyses has allowed structure in the gonococcal population to be identified.
Image