Treating gonorrhoea
Treatment is indicated following a positive identification of Neisseria gonorrhoeae by nucleic acid amplification testing (NAAT) and/or culture to test for susceptibility and identify resistant strains.
In the UK, gonorrhoea is treated with a single antibiotic injection such as ceftriaxone (1g, an increase from the previously recommended dose of 500 mg) however, if antimicrobial susceptibility test results from all sites of infection are available and, the isolate is sensitive to ciprofloxacin, then this should be used for treatment in preference to ceftriaxone.
Image

Given the rise in resistance to azithromycin, guidelines published in 2019 by the British Association for Sexual Health and HIV (BASHH) no longer recommend its use in dual therapy to treat gonorrhoea. Guidelines also advised extra-genital testing for patients with known or suspected antimicrobial resistance.
With effective treatment, most symptoms should improve within a few days however, all patients should be advised for a follow-up and test of cure (TOC) to ensure the infection has cleared.
Image

The British Association for Sexual Health and HIV (BASHH) provides guidelines on multiple sexually transmitted infections and you can find out more about this here.
The increasing numbers of cases of gonorrhoea globally is concerning because the gonococcus is becoming resistant to antibiotics to the extent that this infection may not be as easy to cure in the future.
This is important as untreated gonorrhoea can lead to significant sexual health problems including infertility and pelvic inflammatory disease. Gonorrhoea in pregnancy also poses considerable risks to the unborn baby.
Most of the genetic determinants known to confer resistance in N. gonorrhoeae are encoded on the chromosome, with plasmid-mediated AMR associated with the blaTEM gene conferring high level resistance to penicillin and the tetM gene associated with resistance to tetracycline.
All of the genes known to confer resistance have been defined in the PubMLST.org/nesseria database and can be annotated in whole genome sequence data. This includes genes conferring resistance to:
Image
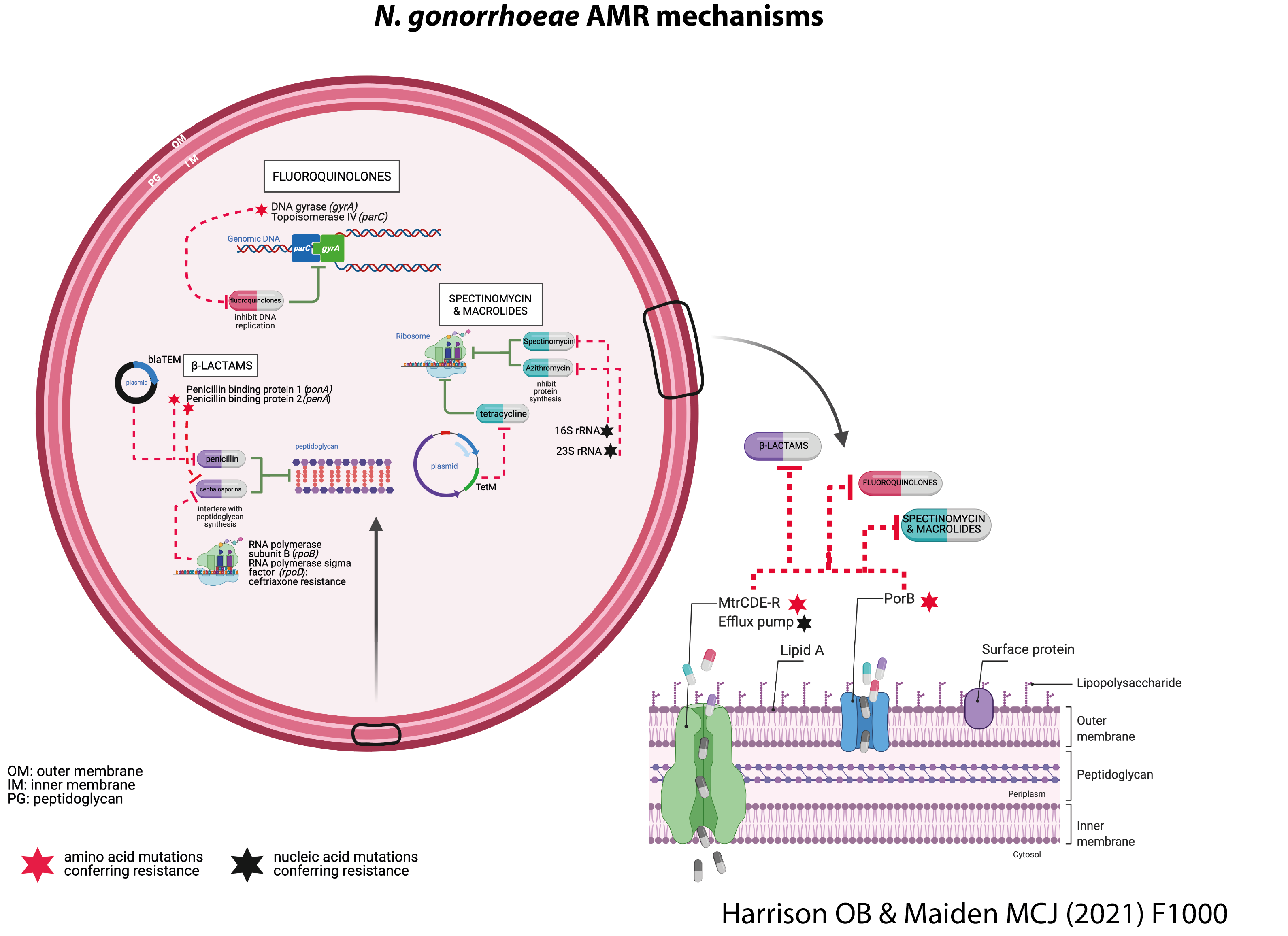
The annotation of genes known to confer resistance in whole genome sequence data are helping us combat antimicrobial resistance. We can, for example, understand how resistance evolves and identify mutations conferring resistance. This knowledge can then be applied to monitor the global distribution of resistant strains.
In particular, we have found an association between certain gonococcal strains and resistance. You can explore these associations using the data below.
In this interactive visualisation, we are examining antibiotic susceptibility tendencies exhibited within a well-defined gonococcal isolate collection. You can find this isolate collection on PubMLST by clicking here.
The far left panel depicts the antibiotics: Ciprofloxacin, azithromycin, cefixime, ceftriaxone, penicillin and tetracycline.
The second from left panel represents core genome groups the gonococci belong to.
The second from right indicates the NG STAR ST and with the far right the country of origin for particular isolates.
You can select from the drop down box specific years, core genome groups and/or antibiotic. By exploring this data you will observe distinct associations between antibiotic phenotype and core genome group.
S: susceptible; I: intermediate resistance; R: resistant. Antibiotic susceptibility profiles derived using breakpoints determined by EUCAST v.11.0