Our Results
Infectious diseases continue to have a major impact on human lives. Our research has contributed to reducing this impact since 1988. We are producing visualisations of data for various diseases.
Why is characterisation and surveillance of Neisseria meningitidis important?
Invasive meningococcal disease (IMD) is a leading cause of bacterial meningitis throughout the world. The geographic distribution and epidemic potential differ according to the type and varies over time. Early public health intervention is essential to save lives and reduce complications.

Phenotype and genotype characterisation are essential to understanding the epidemiological and population biology of the meningococcus. Together these data types allow us to identify related isolates and confirm outbreaks, monitor trends and the distribution of types, detect antibiotic resistance, and estimate the impact of vaccine programs. The Global Meningitis Genome Library brings together these data to explore, examine, and anaylse the population structure of invasive meningococcal disease, and using visual analytics, create new insights.
Which variants of Campylobacter bacteria cause food poisoning?
There are 35 species of Campylobacter, but only two of these cause most human disease.
In the UK, Campylobacter jejuni causes about 90% and Campylobacter coli cause about 10%.

The bacterium Campylobacter causes food poisoning and worse. It is found naturally in the guts of animals we eat and is apparently harmless to them. When it enters the food chain via contamination or we are exposed directly two species of Campylobacter make us sick. MaidenLab sequences the genomes of these bacteria and links this data to cases of gastroenteritis. We have created visual ways for you to look at our data and invite you to explore these and the data too.
How can you use different genomic methods to identify Klebsiella species?
Accurately identifying bacterial species is an essential part of microbiology, with relevance to many scientific disciplines and medical sciences.
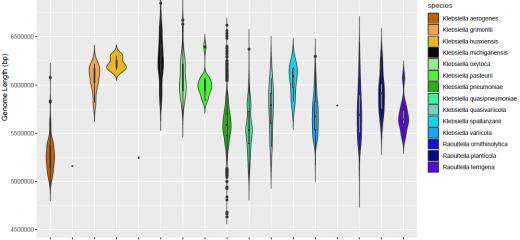
Defining bacterial species is a complex task, involving assessments of phenotypic and genetic characteristics, which are unique to a set of bacteria and differ sufficiently from others. There are many important species within the Klebsiella genus that are of interest to scientists and clinicians worldwide. It is therefore important for them to be able to accurately identify the species from the 13 Klebsiella spp. recognised to date and the 4 closely-related Raoultella spp.
Using Genomics to combat STDs
How can genomics help us?
The WHO estimates that around 1 million new sexually transmitted infections are acquired every day. In England, gonorrhoea and syphilis diagnoses have risen by 277% and 199% since 2010, with a 49% increase in Chlamydia trachomatis diagnoses. The rising trend in STDs is compounded by an increasing prevalence of N. gonorrhoeae and Treponema pallidum subsp. Pallidum strains exhibiting antimicrobial resistance. This worrying trend has galvanised public health authorities worldwide into action leading to improved diagnostic services, increased public awareness of STDs and enhanced surveillance. Our work has begun with the analysis of whole genome sequence data belonging to several thousand gonococcal isolates. Explore our data here to see these data can help us understand gonococcal populations.