What we do
The main focus of MaidenLab is the study of pathogens, the microorganisms (germs) that make people sick. We mostly work on bacterial pathogens, which are very different from viral pathogens, such as influenza or SARS-CoV-2, and ‘parasites’, eukaryotic pathogens such as Plasmodium falciparum, which causes malaria. There are very many bacterial pathogens, which cause diseases as different as tuberculosis and plague. We are especially interested in bacterial meningitis and septicaemia, gastroenteritis, and sexually transmitted diseases (STDs).
Our work starts by considering how disease fits into the life-cycle of the pathogen. By understanding this we then help to design healthcare interventions that reduce the impact of the disease. We use an approach called Population Genomics, which combines sampling the population of the pathogen with the study of the genome of the pathogen. Sampling the population well requires us to understand how the organism lives and how it is distributed in nature. The genome is the instruction set that all organisms contain. Over the last 20 years or so we have learned to read all of this information, leading to the science of genomics. Although we can read most or all of the instructions, or genes, we are still a long way from being able to understand them all. We can achieve understanding by combining the information in the genes in the genome (the genotype) with what a bacterium does (its phenotype) and when and where the organism comes from (its provenance). Over the past 30 years we have constructed a means of undertaking Population Genomics that we call Population Annotation, which aspires to annotate and catalogue the variation of every gene in every organism in a population. We make all of our data and tools widely available on the Internet. Central to these efforts is the PubMLST.org website.
In 1996-1998 we led the development of an approach called multi-locus sequence typing (MLST), which was an early means of linking genotype with phenotype and provenance data. To support this we placed the data on a website. We have moved a long way since then. The current PubMLST.org (for Public MLST) website is capable of hosting complete genome sequences and associated data for hundreds of thousands of bacterial isolates. Analysis tools are included and all the information can be made freely available via the PubMLST.org website or with an automated computer-to-computer data sharing system (Application Programming Interface, or API).
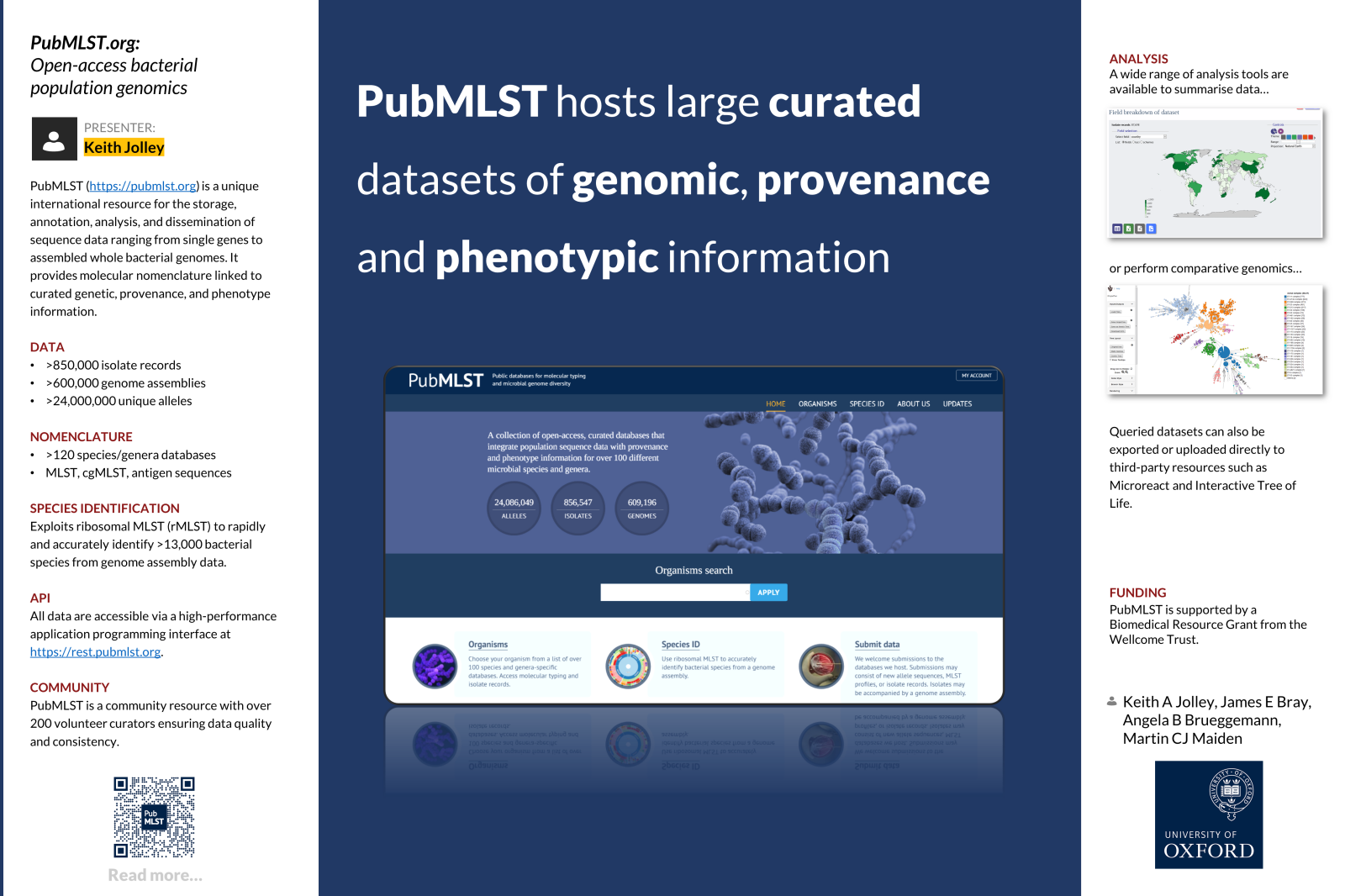
At the heart of PubMLST is the Bacterial Isolate Genome Sequence database (BIGSdb) software. This open-access software enables the storage and organisation of sequence data, diversity data and provenance and phenotype data.
The Pathogenic Neisseria and their relatives
Campylobacter
Bioinformatics and method development
Collaborations